Prioritization and analysis of PIK3CA driver mutations with COSMIC Cancer Mutation Census

Prioritization and analysis of PIK3CA driver mutations with COSMIC Cancer Mutation Census
You can read the original post here
COSMIC (Catalogue Of Somatic Mutation In Cancer) is the world’s largest, most comprehensive database of somatic mutation information relating to human cancer. Over the course of more than 20 years, COSMIC has expanded to include a broad range of manually expertly curated datasets and products. These aim to support scientific understanding of the impact of somatic mutations in cancer and facilitate improvements in the field of precision oncology. In 2020, COSMIC released the Cancer Mutation Census (CMC) and here, we guide you through what CMC has to offer and how these insights can boost your research.
What is the Cancer Mutation Census (CMC)?
The Cancer Mutation Census, first launched in 2020, as part of the wider COSMIC knowledge base.
CMC is a dataset and webtool within COSMIC aimed at identifying coding mutations with a potential to drive cancer. This is achieved by combining manually curated information regarding cancer genes and genetic variants with data on variant frequencies in cancer and non-cancer populations, and algorithmic evaluation of variant significance. It applies a simple and transparent set of rules to the whole set of coding mutations in COSMIC to identify variants with the highest evidence of functional significance and clinical relevance. The computational assessment of each mutation takes into account a range of variables including ClinVar significance, dN/dS ratios, variant frequencies in normal populations (gnomAD) and more, and assigns each mutation into one of four possible tiers of confidence.
Tier 1 is the highest confidence set, tiers 2 and 3 contain variants with significant, but less extensive evidence of involvement in carcinogenesis. Finally, mutations with little to no evidence for driving cancer are classified as Tier 4. In addition to this classification, CMC integrates and presents the information used to prioritise variants, including their frequencies in various cancer types (COSMIC), germline frequencies (gnomAD), ClinVar annotations, dN/dS analysis results, and nucleotide and amino acid conservation.
PIK3CA spotlight
PIK3CA as an oncogene
PIK3CA is an oncogene frequently mutated or amplified in a wide range of cancers, it encodes the catalytic subunit of PI3K, which is active in the signal transduction of several receptor tyrosine kinase (RTK) signalling pathways that control a range of cellular activities. PIK3CA has been shown to promote several cancer hallmarks such as proliferation migration, angiogenesis and survival.
Clinical significance
A Tier 1 Cancer Gene Census gene, PIK3CA is well documented as an oncogenic driver of cancer. The prevalence of PIK3CA mutations in a range of cancers including breast, colorectal, urinary tract and endometrial cancer clearly indicates that a comprehensive understanding of the clinical relevance of these mutations is imperative. This gene and its associated mutations are highly clinically relevant, predictive of prognosis and response to drugs in different cancer types and thus accurate and efficient prioritisation of these potential therapeutic targets is crucial.
CMC data
The Cancer Mutation Census currently catalogues 1292 coding mutations for PIK3CA, comprising 952 missense, 141 synonymous, 76 in-frame indels, 53 frameshifts, and 60 truncating mutations. To elucidate the potential implications of these variants, the CMC classifies them into three tiers based on their likelihood of being cancer drivers. Lower tiers indicate a greater probability of a mutation being a cancer driver. This tiering system is based on the fulfilment of three criteria in varying combinations: (1) high recurrence in Cancer Gene Census (CGC) genes, (2) clinical significance (pathogenic or likely pathogenic) as per ClinVar, and (3) evidence of positive selection determined by the dN/dS method.
Tier 1: Mutations that are highly recurrent in a CGC gene and also classified as pathogenic or likely pathogenic in ClinVar.
Tier 2: Mutations that meet any two of the three criteria.
Tier 3: Mutations that meet one of the criteria.
Unclassified (Others): Mutations that do not meet any of the criteria.
For PIK3CA, there are 20 Tier 1 mutations, 23 Tier 2 mutations, and 46 Tier 3 mutations. This tiered classification serves as a foundational step in prioritising mutations in cancer driver genes. By delving into these individual mutations and the bioinformatic analyses aggregated within the CMC, the clinical relevance and potential negative consequences of these mutations become apparent.
After exploring CMC data on this mutation, you can then begin to explore other COSMIC datasets to understand the impact of mutation on function.
CMC and Interoperability
Structural consequences of variants
A feature COSMIC prides itself on is the interoperability of our datasets which collate to provide a comprehensive overview, whether you begin studying from a gene, tissue or mutation perspective. As an example, we can use COSMIC 3D to further explore the consequence of the PIK3CA mutation, p.H1047R which can be found in Tier 1 of the CMC.
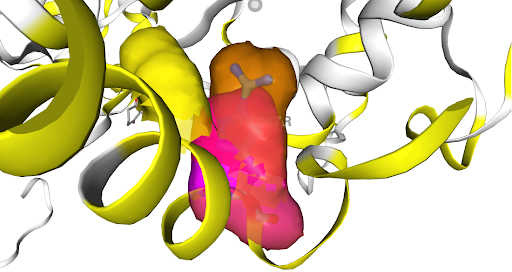
Figure 1: Structural consequence of the p.H1047R mutation shown on a 3D protein model of the PIK3CA gene (Via COSMIC 3D)
Not only can you view the structural consequences of the mutation (as shown above), COSMIC 3D delves into the phenotypic result of this change. In the context of the PIK3CA gene, the p.H1047R mutation is a well-known activating mutation in the catalytic subunit of phosphatidylinositol 3-kinase (PI3K). This mutation is associated with various cancers because it leads to increased kinase activity, promoting cell growth, survival, and proliferation. The specific alteration from histidine to arginine at position 1047 enhances the enzyme's activity, contributing to oncogenic signalling pathways.
COSMIC 3D also enables users to identify predicted small molecule binding sites and evaluate their potential for 'druggability'—the likelihood that these sites could be effective targets for precision oncology drugs. The p.H1047R mutation is considered highly druggable, with a predicted druggability score of 0.79. This feature highlights its potential as a target for therapeutic intervention in cancer treatment
Application in precision oncology
Once the impact of a mutation on protein function and its druggability are established, users can explore these targets within the framework of precision oncology. Via COSMIC Actionability, users can determine whether there are drugs available that target either the protein or a specific mutation, whether these drugs are in clinical trials, or if they have already been approved and are in use.
For instance, the p.H1047R mutation in the PIK3CA gene has been identified as potentially pathogenic and linked to multiple cancer types, as supported by data from the CMC. To begin investigating their mutation of interest, users can filter the Actionability dataset by various parameters such as gene, mutation, and tissue type. By selecting PIK3CA and the p.H1047R variant, it becomes apparent that no marketed drugs with proven efficacy against this mutation have been approved by the FDA or other regulatory authorities. However, there are currently 12 clinical trials in progress, with 4 in the second phase. These trials focus on endometrial tissues, neoplasms, and carcinomas, representing only a small subset of the cancers that this variant may drive.
This highlights a gap in testing of the currently available treatments and underscores the need for further investigation into driver mutations within a critical cancer gene like PIK3CA. Addressing this gap opens the door for potential precision drug approval opportunities, emphasising the importance of targeted therapeutic strategies in combating cancer.
The main takeaway
COSMIC’s Cancer Mutation Census encompasses much more than a simple list of mutations in human cancer. By integrating information from a plethora of trusted sources, the CMC offers gold-standard data for benchmarking algorithms that predict driver and passenger mutations, benefiting from the rigorous manual curation provided by COSMIC. This tool not only supports a more efficient identification of known and predicted driver mutations, but also provides a means to confidently select cancer driver mutations that have been curated and scored with a high degree of confidence for downstream research and development pipelines in pharmaceutics, diagnostics and beyond. Identifying gaps within research, and prioritising only the most imperative targets allows the cancer research community to leave no stone unturned.



