New data integration and annotation system: genomic coordinates for each mutation
To ensure standardisation and interoperability across the entire dataset, the COSMIC database has undergone an extensive update, which has substantially improved the identification of unique variants defined at the genome, transcript, or protein level. We have introduced a new stable genomic Identifier (COSV) that indicates the position of the variant in the genome, allowing variants to be mapped between human genome assemblies, as well as between various transcriptome assemblies. The new identifier has replaced previously used transcript-level (COSM) and non-coding genomic (COSN) identifiers. However, to secure backwards data compatibility, the legacy COSM and COSN identifiers are still included in COSMIC variant reporting and interoperable with COSV identifiers.
All the transcripts and proteins are only from the high quality GENCODE basic set (GENCODE v28) (15). To ensure standardisation and uniformity in the dataset, all the variants with known genomic coordinates are mapped across transcripts and genes using the Ensembl's Variant Effect Predictor (VEP) (16), improving compliance with current HGVS standards (12) and interoperability with other resources.
Changes to curation database and new data export files
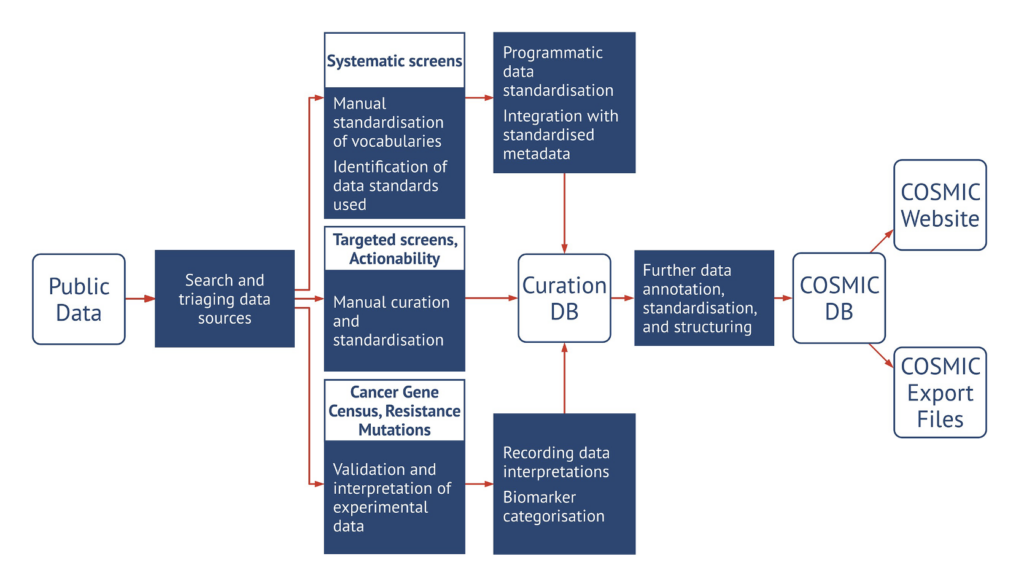
As part of COSMIC’s database restructuring, the curated data was remodelled into a new relational database, to ensure the data is more interconnected, standardised and streamlined. This improved database was used to produce a new set of downloadable export files and download website in v98. The new files both assure improved interoperability of data by making each file more interconnected with internal and external identifiers, and are characterised by reduced data redundancy across all files. All major data points are linked with 10 COSMIC identifiers, which can be used to effectively join the data content from multiple files: COSMIC Phenotype Id (COSO), COSMIC Gene Id (COSG), COSMIC Sample Id (COSS), COSMIC Structural Id (COST), COSMIC CNV Id (COSCNV), COSMIC Gene Fusion Id (COSF), Legacy Mutation Id (COSM/COSN), COSMIC Paper Id (COSP), COSMIC Study Id (COSU), COSMIC Genomic Mutation Id (COSV). Furthermore, each download file is now packaged with a read-me file, while a consistent file name convention helps users to better navigate between the genome assemblies, product releases and to construct their analytic pipelines.
New resources
Mutational Signatures catalogue v3.3 (https://cancer.sanger.ac.uk/signatures/) is a resource developed as part of the CRUK Mutographs Cancer Grand Challenge and a collaborative project involving the Wellcome Sanger Institute, the Pillay lab at University College London, and the Alexandrov lab at the University of California (17). Mutational signatures are patterns of somatic mutations associated with specific mutagenic processes. They can be considered a way of identifying various types of damage within the genome that have accumulated prior to, and during the development and progression, of a cancer. A mutational signature of a cancer can be informative with regard to the nature of the likely source of DNA damage (e.g. UV exposure, smoking, exposure to specific treatments etc.), or be indicative of inactivation of one of the DNA damage repair pathways. For example mutational signatures are used to detect the deficiency of the homologous recombination pathway in cancer samples, indicative for therapy with PARP inhibitors (18).
COSMIC’s mutational signature catalogue has been built through the analysis of the PCAWG dataset (2) using the SigProfiler algorithm (17) and additional curation of selected published studies, where data originate from samples exposed to various DNA-altering factors. Currently, four different mutational signature classes are considered, resulting in a set of 155 individual mutational signatures: SBS (single base substitution signatures; n = 95), DBS (doublet base substitution signatures; n = 11), ID (insertion deletion signatures; n = 18), CN (copy number signatures; n = 21). Within these four classes, each reference signature is described, acknowledging its validation status, proposed aetiology and tissue distribution, potential associations with other mutational signatures and various genomic features, and how the signature has changed compared to the previous versions of the resource.
A recently added feature, the SigProfiler Assignment web tool (19) (https://cancer.sanger.ac.uk/signatures/assignment/) allows users to upload their own sequenced samples in VCF or MAF format to assign the reference mutational signatures to these samples, utilising a custom implementation of the forward stagewise algorithm and non-negative least squares.
The Cancer Mutation Census (https://cancer.sanger.ac.uk/cmc/home) integrates biological, biochemical and population information from multiple sources, allowing users to discover and understand which mutations can drive different types of cancer. It is aimed at improving the application of precision oncology and to help users understand which mutations are likely to be causing cancer, and which are only a result of the disease – passenger mutations.
To assess the oncogenic potential of each of over 5 million coding mutations in COSMIC, manually curated information regarding cancer genes obtained from the Cancer Gene Census and genetic variants, acquired from ClinVar (20) is integrated with data on variant frequencies across multiple cancer types from the COSMIC catalogue. For each variant, this is supported by information on its germline frequency in normal populations (gnomAD) (21), signs of positive selection in cancer cells (dN/dS) (22), as well as conservation on DNA (GERP) (23) and protein (SIFT) (24) level. A simple and transparent set of rules is applied to identify variants with the highest potential of clinical relevance. The CMC website (https://cancer.sanger.ac.uk/cmc) presents the distribution of all CMC mutations, their characteristics and properties within each gene helping to identify clusters of significant mutations, presence and distribution of SNPs in that gene, or assessing conservation of certain fragments of the gene compared to others.
All the data in the CMC are downloadable and interpretations are fully transparent to provide clarity as to why a mutation is classified with either high or low impact. Users are able to see the biological reasoning and the evidence base used.
The scope of COSMIC Actionability is all interventional oncology clinical trials or case studies that have either selected patients by the presence of a variant, or state the intention to correlate efficacy with the presence of a variant. Studies that require the patient to express a protein target or compare efficacy between patients expressing at differing levels are also included. To date (actionability v10), clinical data has been curated for 988 actionable variants, including 156 point mutations, in 445 genes. A total of 11121 trials are represented with 1873 treatments, in 5174 treatment combinations across 226 cancer types.
All Actionability records indicate the source of the data and aim to represent the most complete and recent trial results. As clinical trials rarely identify variants at the DNA level, variants in Actionability are identified at the protein level. Actionability content is curated by gene; a gene is considered fully curated when every relevant trial identified by systematic manual searching in clinical trial repositories and the scientific literature has been recorded. Content for fully curated genes is updated at every release and all previously-recorded trials are re-examined for results updates and new trials, initiated since the previous release.
Resources with updated and expanded content
The Cancer Gene Census (CGC) (7) (https://cancer.sanger.ac.uk/census) is a catalogue of genes affected by somatic alteration and/or rare pathogenic germline mutation that contribute to cancer development. The CGC is compiled by manual curation of peer-reviewed, published mutation data and experimental evidence demonstrating that a gene has a function relevant to cancer. For each gene, the CGC details the cancer types most frequently affected by somatic alteration and/or rare pathogenic germline mutation, the types of somatic alteration affecting the gene in cancer, and the role, or roles, of the gene in one, or more, cancer types. In COSMIC release v98, the CGC includes 738 genes (compared to 719 in v86), partitioned into two tiers (Tier 1: 579 genes; Tier 2: 159 genes) based on the strength of the evidence that associate them with cancer. Each gene is manually evaluated for the presence of somatic mutation patterns typical for cancer drivers (e.g. hotspots of missense or inframe indel mutations characterising oncogenes) and evidence of molecular function associated with potential to drive hallmarks of cancer. For a gene to be qualified to Tier 1, both types of evidence must be present in at least two independent sources. Tier 2 of the CGC comprises genes with convincing evidence of their involvement in cancer development that do not fully fulfil all the requirements for Tier 1 qualification.
In COSMIC release v98, Hallmarks of Cancer annotations are provided for 352 out of 579 CGC Tier 1 genes (92 additional genes characterised since v86). The Hallmarks of Cancer are the phenotypic characteristics shared by cancers (25–27). In COSMIC, these disease-level traits have been adapted to provide a means of describing how genes functionally contribute to cancer development. To achieve that, available publications are studied by a curator to identify gene's involvement in cellular processes that participate in generation of the hallmarks of cancer, such as control of cell division, conducting proliferative signalling, or triggering apoptosis. This often requires performing independent interpretations of experimental results and includes specifying if the function of the protein has potential to drive or suppress each hallmark. Every ‘Hallmark gene page’ provides graphical and tabular summaries of which cancer hallmarks are affected by a wild type protein. Up to 22 data fields collectively constitute a cancer-associated fully referenced mini-gene review. These annotations include summaries of gene function, somatic alterations (including gene fusions) and high penetrance germline mutations that affect the gene in cancer, the impact of coding mutations on protein function, and clinically-relevant gene attributes. New Hallmark gene pages are added to every COSMIC release.
The catalogue of somatic mutations causing drug resistance. COSMIC annotates mutations identified in the literature as conferring resistance to treatment, specifically the mutations acquired after the treatment. In the case of solid tumours, tumours from patients that have received therapy and reported a drug response are annotated in COSMIC using a Drug Response Tumour Feature, based on RECIST (Response Evaluation Criteria in Solid Tumours) (28) terms. Acquired resistance mutations occur in tumours annotated as having ‘resistant recurrence’ drug response phrase. Those acquired mutations reported to be associated with resistance, or presumed by authors to be associated with resistance, are annotated as secondary resistance mutations and included in COSMIC’s catalogue.
Through the export files COSMIC v98 provides information curated from 2945 patient samples, describing 476 mutations in 39 genes responsible for secondary resistance to 60 drugs in 111 cancer types.
The Cancer Browser (https://cancer.sanger.ac.uk/cosmic/browse/tissue) allows website users to query the database from a disease-specific start point, allowing for selection and filtering of COSMIC data by tissue type and histology, i.e. cancer type. Mutation frequencies and profiles within selected disease specific subsets can be explored. The cancer browser lists 49 primary tissue types at the first level of selection in the tissue browser. With subsequent refinements for sub-tissue, histology and sub-histology, the browser provides insights into the somatic alterations of 4161 cancer types. The most frequently mutated genes are graphically displayed for the selected subset. Each gene can then be further explored by displaying its mutation profile within that subset.
COSMIC-3D (29) (https://cancer.sanger.ac.uk/cosmic3d) integrates cancer mutations with protein structure data across the human genome and structural proteome, showcasing over 53 000 experimentally determined protein structures with interactive 3D visualisations to help understand the impact of somatic mutations in the context of protein 3D structure. The resource is updated with new mutations and newly available PDB protein structures at every COSMIC release.
Access to COSMIC
Websites
All COSMIC resources are accessible through the main COSMIC web portal (https://cancer.sanger.ac.uk), which offers summaries and visualisations of data, providing gene profiles including mutation frequencies across 4161 cancer types, detailed nature of mutations, and where available, functional consequence of gene dysfunction via Hallmarks of Cancer, gene expression (z scores) and methylation data. The likely functional significance and clinical actionability of specific mutations can be explored separately through the Cancer Mutation Census and Actionability websites, respectively.
Downloads
For power users, the full content of the COSMIC knowledgebase is available through a collection of export files. Data from the core COSMIC database, the Cancer Gene Census, Cancer Mutation Census, Cell Lines Project, Actionability, as well as definitions of mutational signatures are all available for download; as COSMIC-3D represents data from COSMIC in new visual forms, no downloadable content is provided. All data referring to a human genome are available for both GRCh37 and GRCh38 genome assemblies. The core coding mutation content is available in a tabular form as well as files in vcf format, and other types of genetic alterations, such as structural variants, non-coding variants, gene fusions, gene expression variants, differential DNA methylation, or resistance mutations are provided within specific files.
Genome wide datasets for download can be found on the download page. https://cancer.sanger.ac.uk/cosmic/download for COSMIC, Cancer Gene Census, Cancer Mutation Census, Cell Lines Project, and Actionability data, or https://cancer.sanger.ac.uk/signatures/downloads/ for the mutational signatures’ definitions.
Licensing
To download COSMIC data and use certain functionalities of the websites (CMC), users must register for a COSMIC account. COSMIC is available for free to non-commercial users, whilst a licence fee is applicable to commercial users to support continued growth and maintenance of the resource. Both commercial and non-commercial users have full access to all the data for each new COSMIC release.
Summary
COSMIC’s role in the global cancer research ecosystem is helping the research community conduct their science better and faster by delivering biologically and clinically-relevant genomic knowledge across all cancer types. Through provision of sustainable and accessible bioinformatic datasets and software COSMIC also aims to bridge academic and patient-specific applied sciences. Integrating, standardising and adding highly curated annotation to the data enhances its value and provides a knowledge base to drive current and future research. As an integral part of the research community, COSMIC actively addresses many common challenges faced by those involved in genomic research. These are important factors that will determine the future development of COSMIC and similar resources. One prominent issue is the lack of diversity in genomic datasets. Differences in variant prevalence between global populations are a well-known phenomenon in the germline variant research (30) but in certain cases also patterns of somatic mutations differ between patients with the same cancer type but different ethnic origin (31). Translating these genetic differences into effective clinical practice requires more comprehensive and unbiased genomic datasets. Therefore, an effort has to be made to maximise ethnic and phenotypic diversity in public datasets. Additionally, the absence of widely accepted standards for reporting, processing, and sharing genomic data hinders valuable scientific and clinical information from entering the public domain. These challenges, however, are all addressable. The COSMIC team remains committed to collaborating with scientists and clinicians to overcome these obstacles, thereby fostering the continued development of resources like COSMIC for the betterment of cancer research and patient care.